Graphs provide us with a very useful data structure. They can help us to find structure within our data. With the advent of Machine learning and big data we need to get as much information as possible about our data. Learning a little bit of graph theory can certainly help us with that.
Here is a Graph Analytics for Big Data course on Coursera by UCSanDiego which I highly recommend to learn the basics of graph theory. You can start for free with the 7-day Free Trial.
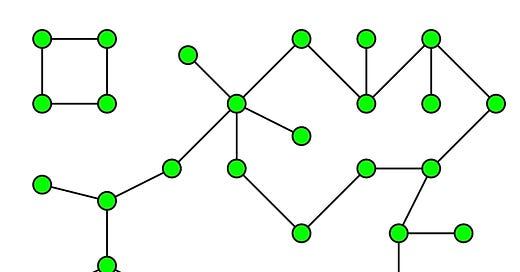
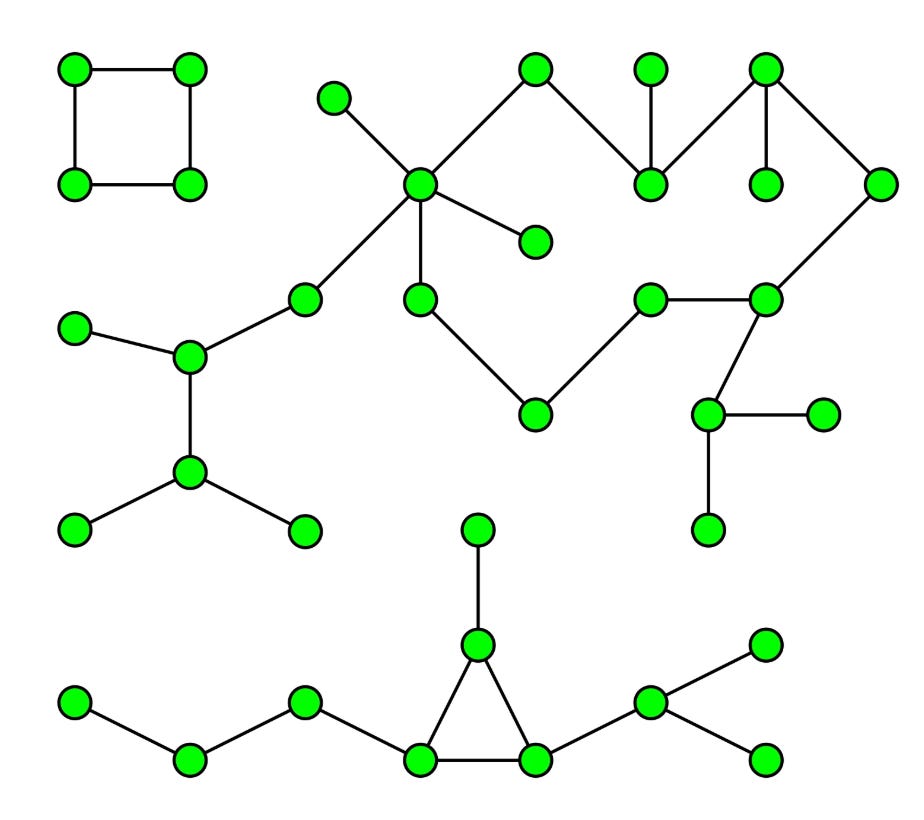
One of the algorithms I am going to focus in the current post is called Connected Components. Why it is important. We all know clustering.
You can think of Connected Components in very layman’s terms as sort of a hard clustering algorithm which finds clusters/islands in related/connected data. As a concrete example: Say you have data about roads joining any two cities in the world. And you need to find out all the continents in the world and which city they contain.
How will you achieve that? Come on give some thought.
To put …
Keep reading with a 7-day free trial
Subscribe to MLWhiz | AI Unwrapped to keep reading this post and get 7 days of free access to the full post archives.